Generate Micro Benchmarks for Velox Backend
This document explains how to use the existing micro benchmark template in Gluten Cpp.
A micro benchmark for Velox backend is provided in Gluten Cpp to simulate the execution of a first or middle stage in Spark. It serves as a more convenient alternative to debug in Gluten Cpp comparing with directly debugging in a Spark job. Developers can use it to create their own workloads, debug in native process, profile the hotspot and do optimizations.
To simulate a first stage, you need to dump the Substrait plan and input split info into two JSON files. The input URIs of the splits should be exising file locations, which can be either local or HDFS paths.
To simulate a middle stage, in addition to the JSON file, you also need to save the input data of this stage into Parquet files. The benchmark will load the data into Arrow format, then add Arrow2Velox to feed the data into Velox pipeline to reproduce the reducer stage. Shuffle exchange is not included.
Please refer to the sections below to learn how to dump the Substrait plan and create the input data files.
Try the example
To run a micro benchmark, user should provide one file that contains the Substrait plan in JSON format, and optional one or more input data files in parquet format. The commands below help to generate example input files:
cd /path/to/gluten/
./dev/buildbundle-veloxbe.sh --build_tests=ON --build_benchmarks=ON
# Run test to generate input data files. If you are using spark 3.3, replace -Pspark-3.2 with -Pspark-3.3
mvn test -Pspark-3.2 -Pbackends-velox -Prss -pl backends-velox -am \
-DtagsToInclude="io.glutenproject.tags.GenerateExample" -Dtest=none -DfailIfNoTests=false -Darrow.version=11.0.0-gluten -Dexec.skip
The generated example files are placed in gluten/backends-velox:
$ tree gluten/backends-velox/generated-native-benchmark/
gluten/backends-velox/generated-native-benchmark/
├── example.json
├── example_lineitem
│ ├── part-00000-3ec19189-d20e-4240-85ae-88631d46b612-c000.snappy.parquet
│ └── _SUCCESS
└── example_orders
├── part-00000-1e66fb98-4dd6-47a6-8679-8625dbc437ee-c000.snappy.parquet
└── _SUCCESS
Run micro benchmark with the generated files as input. You need to specify the absolute path to the input files:
cd /path/to/gluten/cpp/build/velox/benchmarks
./generic_benchmark \
--plan /home/sparkuser/github/oap-project/gluten/backends-velox/generated-native-benchmark/example.json \
--data /home/sparkuser/github/oap-project/gluten/backends-velox/generated-native-benchmark/example_orders/part-00000-1e66fb98-4dd6-47a6-8679-8625dbc437ee-c000.snappy.parquet,\
/home/sparkuser/github/oap-project/gluten/backends-velox/generated-native-benchmark/example_lineitem/part-00000-3ec19189-d20e-4240-85ae-88631d46b612-c000.snappy.parquet \
--threads 1 --iterations 1 --noprint-result --benchmark_filter=InputFromBatchStream
The output should be like:
2022-11-18T16:49:56+08:00
Running ./generic_benchmark
Run on (192 X 3800 MHz CPU s)
CPU Caches:
L1 Data 48 KiB (x96)
L1 Instruction 32 KiB (x96)
L2 Unified 2048 KiB (x96)
L3 Unified 99840 KiB (x2)
Load Average: 0.28, 1.17, 1.59
***WARNING*** CPU scaling is enabled, the benchmark real time measurements may be noisy and will incur extra overhead.
-- Project[expressions: (n3_0:BIGINT, ROW["n1_0"]), (n3_1:VARCHAR, ROW["n1_1"])] -> n3_0:BIGINT, n3_1:VARCHAR
Output: 535 rows (65.81KB, 1 batches), Cpu time: 36.33us, Blocked wall time: 0ns, Peak memory: 1.00MB, Memory allocations: 3, Threads: 1
queuedWallNanos sum: 2.00us, count: 2, min: 0ns, max: 2.00us
-- HashJoin[RIGHT SEMI (FILTER) n0_0=n1_0] -> n1_0:BIGINT, n1_1:VARCHAR
Output: 535 rows (65.81KB, 1 batches), Cpu time: 191.56us, Blocked wall time: 0ns, Peak memory: 2.00MB, Memory allocations: 8
HashBuild: Input: 582 rows (16.45KB, 1 batches), Output: 0 rows (0B, 0 batches), Cpu time: 1.84us, Blocked wall time: 0ns, Peak memory: 1.00MB, Memory allocations: 3, Threads: 1
distinctKey0 sum: 583, count: 1, min: 583, max: 583
queuedWallNanos sum: 0ns, count: 1, min: 0ns, max: 0ns
rangeKey0 sum: 59748, count: 1, min: 59748, max: 59748
HashProbe: Input: 37897 rows (296.07KB, 1 batches), Output: 535 rows (65.81KB, 1 batches), Cpu time: 189.71us, Blocked wall time: 0ns, Peak memory: 1.00MB, Memory allocations: 5, Threads: 1
queuedWallNanos sum: 0ns, count: 1, min: 0ns, max: 0ns
-- ArrowStream[] -> n0_0:BIGINT
Input: 0 rows (0B, 0 batches), Output: 37897 rows (296.07KB, 1 batches), Cpu time: 1.29ms, Blocked wall time: 0ns, Peak memory: 0B, Memory allocations: 0, Threads: 1
-- ArrowStream[] -> n1_0:BIGINT, n1_1:VARCHAR
Input: 0 rows (0B, 0 batches), Output: 582 rows (16.45KB, 1 batches), Cpu time: 894.22us, Blocked wall time: 0ns, Peak memory: 0B, Memory allocations: 0, Threads: 1
-----------------------------------------------------------------------------------------------------------------------------
Benchmark Time CPU Iterations UserCounters...
-----------------------------------------------------------------------------------------------------------------------------
InputFromBatchVector/iterations:1/process_time/real_time/threads:1 41304520 ns 23740340 ns 1 collect_batch_time=34.7812M elapsed_time=41.3113M
Generate Substrait plan and input for any query
First, build Gluten with --build_benchmarks=ON.
cd /path/to/gluten/
./dev/buildbundle-veloxbe.sh --build_benchmarks=ON
# For debugging purpose, rebuild Gluten with build type `Debug`.
./dev/buildbundle-veloxbe.sh --build_benchmarks=ON --build_type=Debug
First, get the Stage Id from spark UI for the stage you want to simulate. And then re-run the query with below configurations to dump the inputs to micro benchmark.
| Parameters | Description | Recommend Setting |
|---|---|---|
| spark.gluten.sql.benchmark_task.stageId | Spark task stage id | target stage id |
| spark.gluten.sql.benchmark_task.partitionId | Spark task partition id, default value -1 means all the partition of this stage | 0 |
| spark.gluten.sql.benchmark_task.taskId | If not specify partition id, use spark task attempt id, default value -1 means all the partition of this stage | target task attemp id |
| spark.gluten.saveDir | Directory to save the inputs to micro benchmark, should exist and be empty. | /path/to/saveDir |
Check the files in spark.gluten.saveDir. If the simulated stage is a first stage, you will get 3 types of dumped file:
- Configuration file: INI formatted, file name
conf_[stageId]_[partitionId].ini. Contains the configurations to init Velox backend and runtime session. - Plan file: JSON formatted, file name
plan_[stageId]_[partitionId].json. Contains the substrait plan to the stage, without input file splits. - Split file: JSON formatted, file name
split_[stageId]_[partitionId]_[splitIndex].json. There can be more than one split file in a first stage task. Contains the substrait plan piece to the input file splits.
Run benchmark. By default, the result will be printed to stdout. You can use --noprint-result to suppress this output.
Sample command:
cd /path/to/gluten/cpp/build/velox/benchmarks
./generic_benchmark \
--conf /absolute_path/to/conf_[stageId]_[partitionId].ini \
--plan /absolute_path/to/plan_[stageId]_[partitionId].json \
--split /absolut_path/to/split_[stageId]_[partitionId]_0.parquet,/absolut_path/to/split_[stageId]_[partitionId]_1.parquet \
--threads 1 --noprint-result
If the simulated stage is a middle stage, you will get 3 types of dumped file:
- Configuration file: INI formatted, file name
conf_[stageId]_[partitionId].ini. Contains the configurations to init Velox backend and runtime session. - Plan file: JSON formatted, file name
plan_[stageId]_[partitionId].json. Contains the substrait plan to the stage. - Data file: Parquet formatted, file name
data_[stageId]_[partitionId]_[iteratorIndex].json. There can be more than one input data file in a middle stage task. The input data files of a middle stage will be loaded as iterators to serve as the inputs for the pipeline:
"localFiles": {
"items": [
{
"uriFile": "iterator:0"
}
]
}
Sample command:
cd /path/to/gluten/cpp/build/velox/benchmarks
./generic_benchmark \
--conf /absolute_path/to/conf_[stageId]_[partitionId].ini \
--plan /absolute_path/to/plan_[stageId]_[partitionId].json \
--data /absolut_path/to/data_[stageId]_[partitionId]_0.parquet,/absolut_path/to/data_[stageId]_[partitionId]_1.parquet \
--threads 1 --noprint-result
For some complex queries, stageId may cannot represent the Substrait plan input, please get the taskId from spark UI, and get your target parquet from saveDir.
In this example, only one partition input with partition id 2, taskId is 36, iterator length is 2.
cd /path/to/gluten/cpp/build/velox/benchmarks
./generic_benchmark \
--plan /absolute_path/to/complex_plan.json \
--data /absolute_path/to/data_36_2_0.parquet,/absolute_path/to/data_36_2_1.parquet \
--threads 1 --noprint-result
Save ouput to parquet to analyze
You can save the output to a parquet file to analyze.
cd /path/to/gluten/cpp/build/velox/benchmarks
./generic_benchmark \
--plan /absolute_path/to/plan.json \
--data /absolute_path/to/data.parquet
--threads 1 --noprint-result --write-file=/absolute_path/to/result.parquet
Add shuffle write process
You can add the shuffle write process at the end of this stage. Note that this will ignore the --write-file option.
cd /path/to/gluten/cpp/build/velox/benchmarks
./generic_benchmark \
--plan /absolute_path/to/plan.json \
--split /absolute_path/to/split.json \
--threads 1 --noprint-result --with-shuffle
By default, the compression codec for shuffle outputs is LZ4. You can switch to other codecs by adding one of the following argument flags to the command:
- –zstd: ZSTD codec, compression level 1
- –qat-gzip: QAT GZIP codec, compression level 1
- –qat-zstd: QAT ZSTD codec, compression level 1
- –iaa-gzip: IAA GZIP codec, compression level 1
Note using QAT or IAA codec requires Gluten cpp is built with these features. Please check the corresponding section in Velox document first for how to setup, build and enable these features in Gluten. For QAT support, please check Intel® QuickAssist Technology (QAT) support. For IAA support, please check Intel® In-memory Analytics Accelerator (IAA/IAX) support
Simulate Spark with multiple processes and threads
You can use below command to launch several processes and threads to simulate parallel execution on Spark. Each thread in the same process will be pinned to the core number starting from --cpu.
Suppose running on a baremetal machine with 48C, 2-socket, HT-on, launching below command will utilize all vcores.
processes=24 # Same value of spark.executor.instances
threads=8 # Same value of spark.executor.cores
for ((i=0; i<${processes}; i++)); do
./generic_benchmark --plan /path/to/plan.json --split /path/to/split.json --noprint-result --threads $threads --cpu $((i*threads)) &
done
If you want to add the shuffle write process, you can specify multiple direcotries by setting environment variable GLUTEN_SPARK_LOCAL_DIRS to a comma-separated string for shuffle write to spread the I/O pressure to multiple disks.
mkdir -p {/data1,/data2,/data3}/tmp # Make sure each directory has been already created.
export GLUTEN_SPARK_LOCAL_DIRS=/data1/tmp,/data2/tmp,/data3/tmp
processes=24 # Same value of spark.executor.instances
threads=8 # Same value of spark.executor.cores
for ((i=0; i<${processes}; i++)); do
./generic_benchmark --plan /path/to/plan.json --split /path/to/split.json --noprint-result --with-shuffle --threads $threads --cpu $((i*threads)) &
done
Run Examples
We also provide some example inputs in cpp/velox/benchmarks/data. E.g. generic_q5/q5_first_stage_0.json simulates a first-stage in TPCH Q5, which has the the most heaviest table scan. You can follow below steps to run this example.
- Open generic_q5/q5_first_stage_0.json with file editor. Search for
"uriFile": "LINEITEM"and replaceLINEITEMwith the URI to one partition file in lineitem. In the next line, replace the number in"length": "..."with the actual file length. Suppose you are using the provided small TPCH table in cpp/velox/benchmarks/data/tpch_sf10m, the replaced JSON should be like:
{
"items": [
{
"uriFile": "file:///path/to/gluten/cpp/velox/benchmarks/data/tpch_sf10m/lineitem/part-00000-6c374e0a-7d76-401b-8458-a8e31f8ab704-c000.snappy.parquet",
"length": "1863237",
"parquet": {}
}
]
}
- Launch multiple processes and multiple threads. Set
GLUTEN_SPARK_LOCAL_DIRSand add –with-shuffle to the command.
mkdir -p {/data1,/data2,/data3}/tmp # Make sure each directory has been already created.
export GLUTEN_SPARK_LOCAL_DIRS=/data1/tmp,/data2/tmp,/data3/tmp
processes=24 # Same value of spark.executor.instances
threads=8 # Same value of spark.executor.cores
for ((i=0; i<${processes}; i++)); do
./generic_benchmark --plan /path/to/gluten/cpp/velox/benchmarks/data/generic_q5/q5_first_stage_0.json --split /path/to/gluten/cpp/velox/benchmarks/data/generic_q5/q5_first_stage_0_split.json --noprint-result --with-shuffle --threads $threads --cpu $((i*threads)) &
done >stdout.log 2>stderr.log
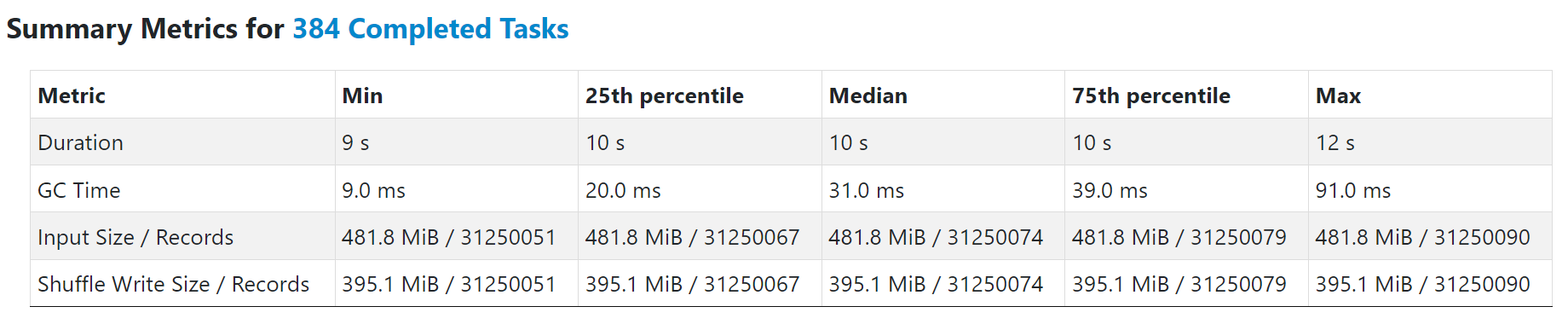
You can find the “elapsed_time” and other metrics in stdout.log. In below output, the “elapsed_time” is ~10.75s. If you run TPCH Q5 with Gluten on Spark, a single task in the same Spark stage should take about the same time.
------------------------------------------------------------------------------------------------------------------
Benchmark Time CPU Iterations UserCounters...
------------------------------------------------------------------------------------------------------------------
SkipInput/iterations:1/process_time/real_time/threads:8 1317255379 ns 10061941861 ns 8 collect_batch_time=0 elapsed_time=10.7563G shuffle_compress_time=4.19964G shuffle_spill_time=0 shuffle_split_time=0 shuffle_write_time=1.91651G